|
|
Recombinant Micrococcal Nuclease
Catalog # : EPX-016-REP
Source : Staphylococcus aureus
Expressed in : E. Coli
Quantity : 500 UN at 0.5U /ul
Background:
Micrococcal nuclease or MNase is a 16.9 kDa endonuclease derived from Staphylococcus aureus (Gene ID: 3238436). It is purified from an E. coli strain expressing an N-terminal 6XHIS tagged micrococcal nuclease.. Purified protein exhibit an strong endonuclease activity against single-stranded, double-stranded, circular and linear nucleic acids. The enzyme is active in the pH range of 7.0 - 10.0, with optimal activity at pH 9.2 for both RNA and DNA substrates. The rate of cleavage is 30 times greater at the 5' side of A or T than at G or C and results in the production of mononucleotides and oligonucleotides with terminal 3’-phosphates
Protein details:
Micrococcal Nuclease (MNase) is suitable for removing nucleic acids from cell lysates, releasing chromatin-bound proteins and shearing chromatin for use in chromatin immunoprecipitation (ChIP) experiments. The enzyme activity is strictly dependent on Ca2+ and the pH optimum varies according to Ca2+ concentration. The enzyme is therefore easily inactivated by EGTA or EDTA. Purified Micrococcal Nuclease is formulated in a storage buffer containing 20mM Tris-Cl pH 8.0, 50mM NaCl, 1mM DTT and 50% glycerol.
Unit Definition:
One unit will produce 1.0 μmole of acid soluble
polynucleotides from native DNA per min at pH 8 at 37 °C, based on
EM/260 = 10,000 for the mixed nucleotides.
Quality control:
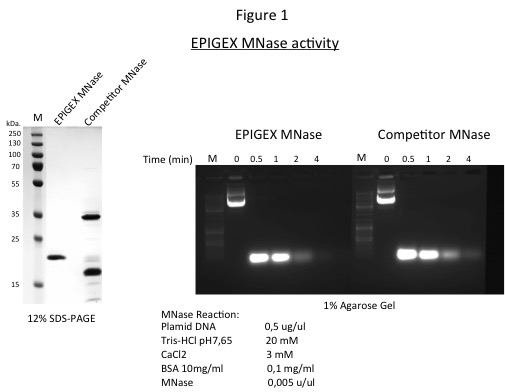
Purified recombinant micrococcal nuclease is evaluated by 12% SDS PAGE mini-gel for protein purity and by spectrometry and 1% agarose gel for nuclease activity (Figure 1, right and left panels).
Application Notes:
For research use only.
Storage:
-20°C
Guarantee:
Products guaranteed stable for 2 years from date of receipt when stored properly.
Purity:
>98% purity by SDS PAGE.
Protein sequences: H2A (Theoretical Mw: 14091.48) MSGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRKGNYAERVGAGA PVYLAAVLEYLTAEILELAGNAARDNKKTRIIPRHLQLAIRNDEELNK LLGKVTIAQGGVLPNIQAVLLPKKTESHHKAKGK
H2B (Theoretical Mw: 13904.17) MPEPAKSAPAPKKGSKKAVTKAQKKDGKKRKRSRKESYSIYVYKVLKQ VHPDTGISSKAMGIMNSFVNDIFERIAGEASRLAHYNKRSTITSREIQ TAVRLLLPGELAKHAVSEGTKAVTKYTSAK
H3.1 (Theoretical Mw: 15404.09) MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRYRPGTVALR EIRRYQKSTELLIRKLPFQRLVREIAQDFKTDLRFQSSAVMALQEACEAY LVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA
H4 (Theoretical Mw: 11367.34) MSGRGKGGKGLGKGGAKRHRKVLRDNIQGITKPAIRRLARRGGVKRIS GLIYEETRGVLKVFLENVIRDAVTYTEHAKRKTVTAMDVVYALKRQGR TLYGFGG
References: 1. Van Holde, K. E. (1989) Chromatin, 1-497. 2. Luger et al., (1997) Nature, 389(6648):251-60.. |
|---|
© 2011-2016 EpiGex. All rights reserved